Flywheel-Connect 3D Slicer Extension

Modern scientific workflows require the utilization of a diverse (and sometimes disparate) set of tools to turn data into insight. With each of these tools designed to excel at a specific set of tasks, shepherding results between tools can require significant effort and expense. Fortunately, Flywheel is an integrated framework facilitating ease-of-use of these tools and the transfer of data between them. The Flywheel-connect tool described here is an 3d Slicer extension which further enhances the Flywheel platform’s integrated functions.
Extending the Flywheel-Connect 3D Slicer Extension
In this article we demonstrate the capacity of Flywheel to extend the utility of 3D Slicer. 3D Slicer is an open source software platform for medical image informatics, image processing, and three-dimensional visualization. While 3D Slicer excels at desktop visualization and local image analysis, it lacks the capacity to orchestrate cloud-centric computing and institutional data resources — the specific domain in which the Flywheel 3D Slicer extension shines.
We have leveraged 3D Slicer’s Extension architecture and Flywheel’s Python SDK to provide 3D Slicer access to NifTI images stored within a remote Flywheel instance running on Google Cloud Platform. Called “flywheel-connect”, this 3D Slicer extension is a proof-of-concept utility for downloading and displaying multiple NifTI images from selected acquisitions within a specific Flywheel instance. The “flywheel-connect” extension makes it possible for a 3D Slicer extension user to directly access and load data stored within any Flywheel instance they have access to.
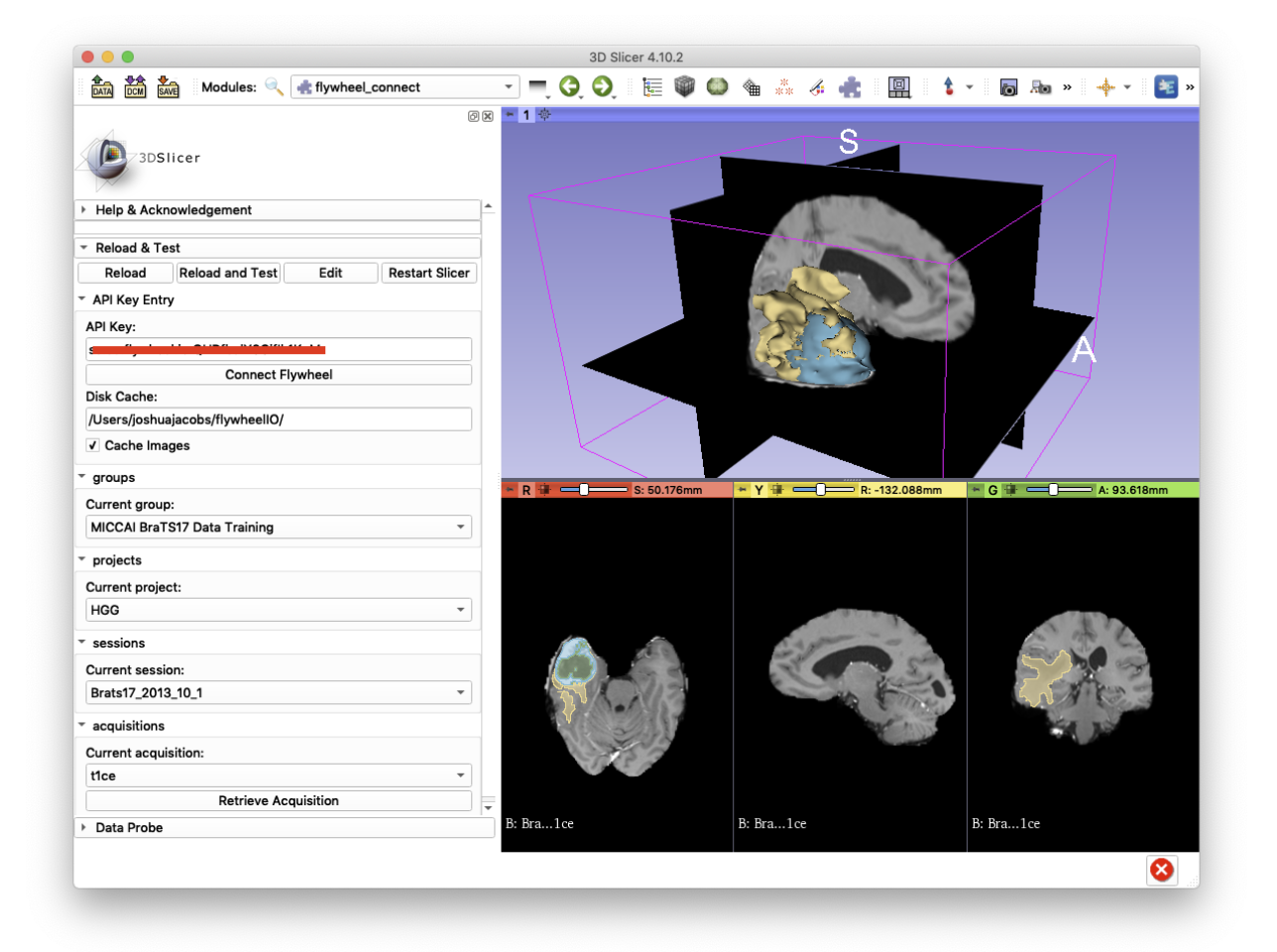
Demonstration
We can demonstrate flywheel-connect below using data from the 2017 MICCAI Multimodal Brain Tumor Segmentation Challenge. Firstly, a user-generated api-key from a Flywheel instance is input. Then, after the “Connect Flywheel” button is pressed, combo-boxes are populated with all of the groups, projects, sessions, and acquisitions that the user has permissions to access. To load an image into the 3D Slicer extension, press “Retrieve Acquisition”. By default, images are cached in a user-specific directory for later use. These directories and images can be deleted directly off of the filesystem or flushed with the “Cache Images” checkbox.
When the data is in 3D Slicer, Slicer-specific operations can be performed on these data. The discrete-valued volume representing tumor segmentation across imaging modalities (FLAIR, T1W, T1CE, T2W) is converted into a label map. In turn, this label map is converted to a three-dimensional representation with Slicer-provided tools.
Although flywheel-connect demonstrates a usable solution, additional features would greatly increase overall utility for Slicer-centric imaging workflows. First, the capacity to view the results of specific Flywheel analyses. This would, for example, allow the visual inspection of image registration to an atlas or template done in Flywheel. Secondly, saving an entire 3D Slicer workspace in a Flywheel instance. Along with instantiating this workspace from a Flywheel instance, this feature would provide functionality entirely lacking in 3D Slicer: Management of multiple workspaces relating to the same project.
To try out flywheel-connect for yourself, see the github page for this project. Suggestions and proposed improvements are welcome. https://github.com/flywheel-apps/flywheel-connect